Key information:
- To verify whether your project falls under CoM and can run jobs on Ascend only, run the following command from your terminal (on Cardinal or Ascend; On Pitzer, you’ll need to load Python first:
module load python/3.9-2022.05), replacing project_code with your actual project account:python /users/PZS0645/support/bin/parentCharge.py project_code - When using CoM project, please add
--partition=nextgenin your job scripts or specify ‘nextgen’ as the partition name with OnDemand apps. Failure to do so will result in your job being rejected. - A list of software available on Ascend can be found here: https://www.osc.edu/content/available_software_list_on_next_gen_ascend
- Always specify the module version when loading software. For example, instead of using module load intel, you must use
module load intel/2021.10.0. Failure to specify the version will result in an error message.
Hardware information:
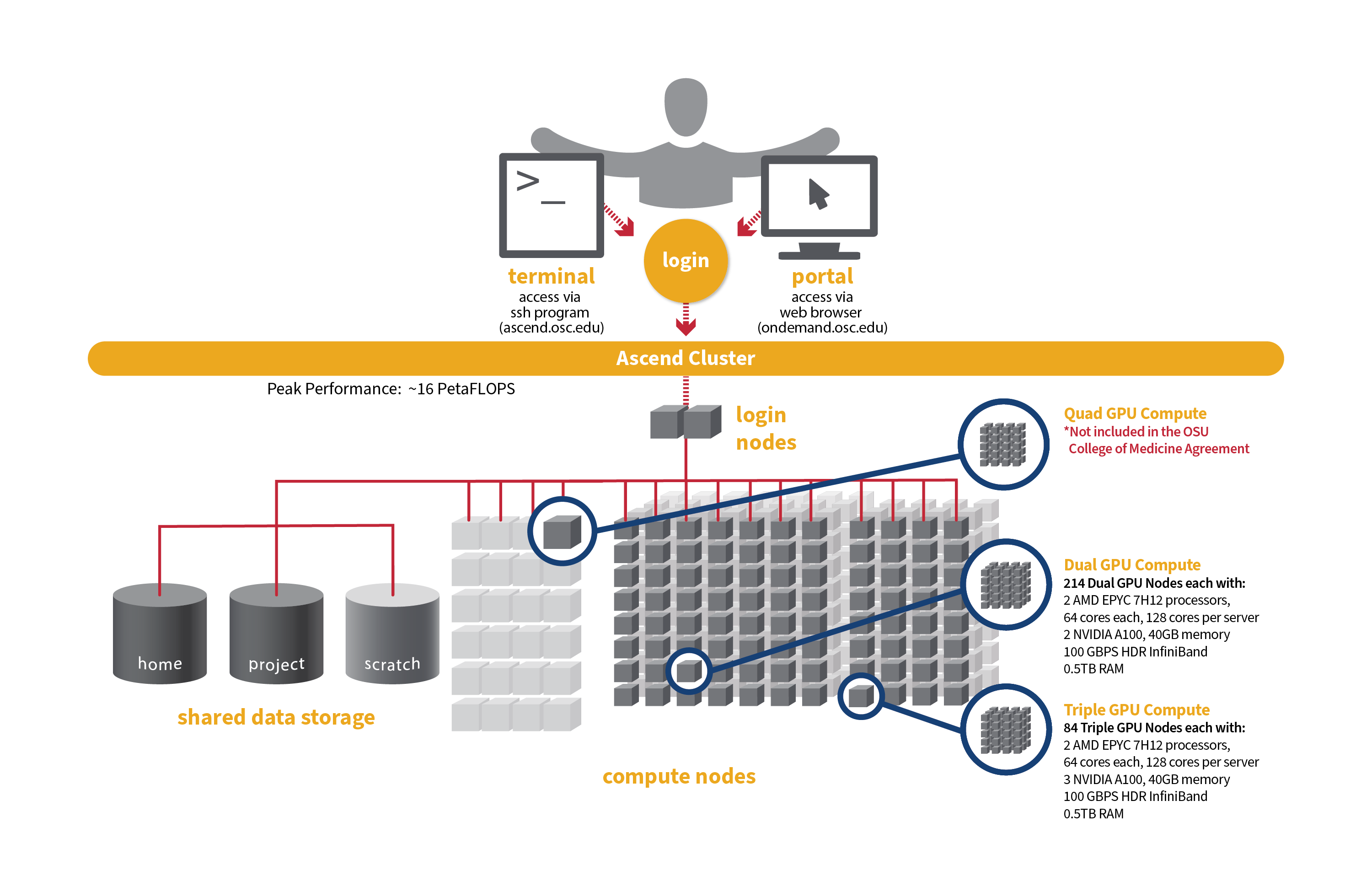
Detailed system specifications for Slurm workload:
- Dual GPU Compute: 190 Dell PowerEdge R7545 two-socket/dual GPU servers, each with:
- 2 AMD EPYC 7H12 processors (2.60 GHz, each with 60 usable cores)
- 2 NVIDIA A100 GPUs with 40GB memory each, PCIe, 250W
- 472GB usable Memory
- 1.92TB NVMe internal storage
- HDR100 Infiniband (100 Gbps)
- Triple GPU Compute: 84 Dell PowerEdge R7545 two-socket/dual GPU servers, each with:
- 2 AMD EPYC 7H12 processors (2.60 GHz, each with 60 usable cores)
- 3 NVIDIA A100 GPUs with 40GB memory each, PCIe, 250W (3rd GPU on each node is under testing and not available for user jobs)
- 472GB usable Memory
- 1.92TB NVMe internal storage
- HDR100 Infiniband (100 Gbps)
Please check this Ascend page for more information on its hardware, programming and software environment, etc.
Governance
The CoM compute service is available to approved CoM users. A regular billing summary for all CoM PIs will be submitted to the OSU CoM Research Computing and Infrastructure Subcommittee (RISST) for review. PIs who are not eligible may be transitioned to a different agreement with OSC.
The committee will also review and consider requests for new project approvals or increases in storage quotas for existing projects.
Storage for CoM projects is billed to CoM at $3.20 per TB/month, with CoM covering up to 10TB. Any additional storage costs may be passed on to the PI.
Set up FY26 budgets
FY26 is the period of July 1, 2025 through June 30, 2026. As a reminder, the project budgets can only be managed by the project PI or a project administrator designated by the PI.
Do the following to create your budget for each project you want to use in FY26:
- Log into MyOSC
- Open the project details
- Select "Create a new budget"
- Select "Add or replace the CURRENT budget" to set the FY26 budget. Use 'unlimited' as the Budget type by choosing 'No' to the question: Do you want to set a dollar budget?
- Confirm your budget dates on the budget review page before submitting
- You will receive an email that your application has been submitted
It may be helpful to review a video explaining how to create and manage budgets.
Creating a new CoM project
Any user with the Primary Investigator (PI) role can request a new project in the client portal. Using the navigation bar, select Project, Create a new project. Fill in the required information.
If you are creating a new academic project
Choose ‘academic’ type as project type. Choose an existing charge account of yours in the College of Medicine, or if you do not have one, create a new charge account and select the department the work will be under. If you cannot find your department, please reach out to us for assistance. Use 'unlimited' as the Budget type by choosing 'No' to the question: Do you want to set a dollar budget?
For more instructions. see Video Tutorial and Projects, budgets and charge accounts page.
If you are creating a new classroom project
Choose ‘classroom’ type as project type. Under the top charge account of CoM: 34754, choose an existing charge account of yours, or if you do not have one, create a new charge account. You will request a $500 budget.
For more instructions. see Video Tutorial and Classroom Project Resource Guide.
Connecting
To access compute resources, you need to log in to Ascend at OSC by connecting to the following hostname:
ascend.osc.edu
You can either use an ssh client application or execute ssh on the command line in a terminal window as follows:
ssh <username>@ascend.osc.edu
From there, you can run programs interactively (only for small and test jobs) or through batch requests.
Running Jobs
OSC clusters are utilizing Slurm for job scheduling and resource management. Slurm , which stands for Simple Linux Utility for Resource Management, is a widely used open-source HPC resource management and scheduling system that originated at Lawrence Livermore National Laboratory. Please refer to this page for instructions on how to prepare and submit Slurm job scripts.
Remember to specify your project codes in the Slurm batch jobs, such that:
#SBATCH --account=PCON0000
where PCON0000 specifies your individual project code.
File Systems
CoM dedicated compute uses the same OSC mass storage environment as our other clusters. Large amounts of project storage is available on our Project storage service. Full details of the storage environment are available in our storage environment guide.
Training and Education Resources
The following are resource guides and select training materials available to OSC users:
- Users new to OSC are encouraged to refer to our New User Resource Guide page and an Introduction to OSC training video.
- A guide to the OSC Client Portal: MyOSC. MySC portal is primarily used for managing users on a project code, such as adding and/or removing users.
- Documentation on using OnDemand web portal can be found here.
- Training materials and tutorial on Unix Basics are here.
- Documentation on the use of the XDMoD tool for viewing job performance can be found here.
- The HOWTO pages, highlighting common activities users perform on our systems, are here.
- A guide on batch processing at OSC is here.
- For specific information about modules and file storage, please see the Batch Execution Environment page.
- Information on Pitzer programming environment can be found here.
Getting Support
Contact OSC Help if you have any other questions, or need other assistance.