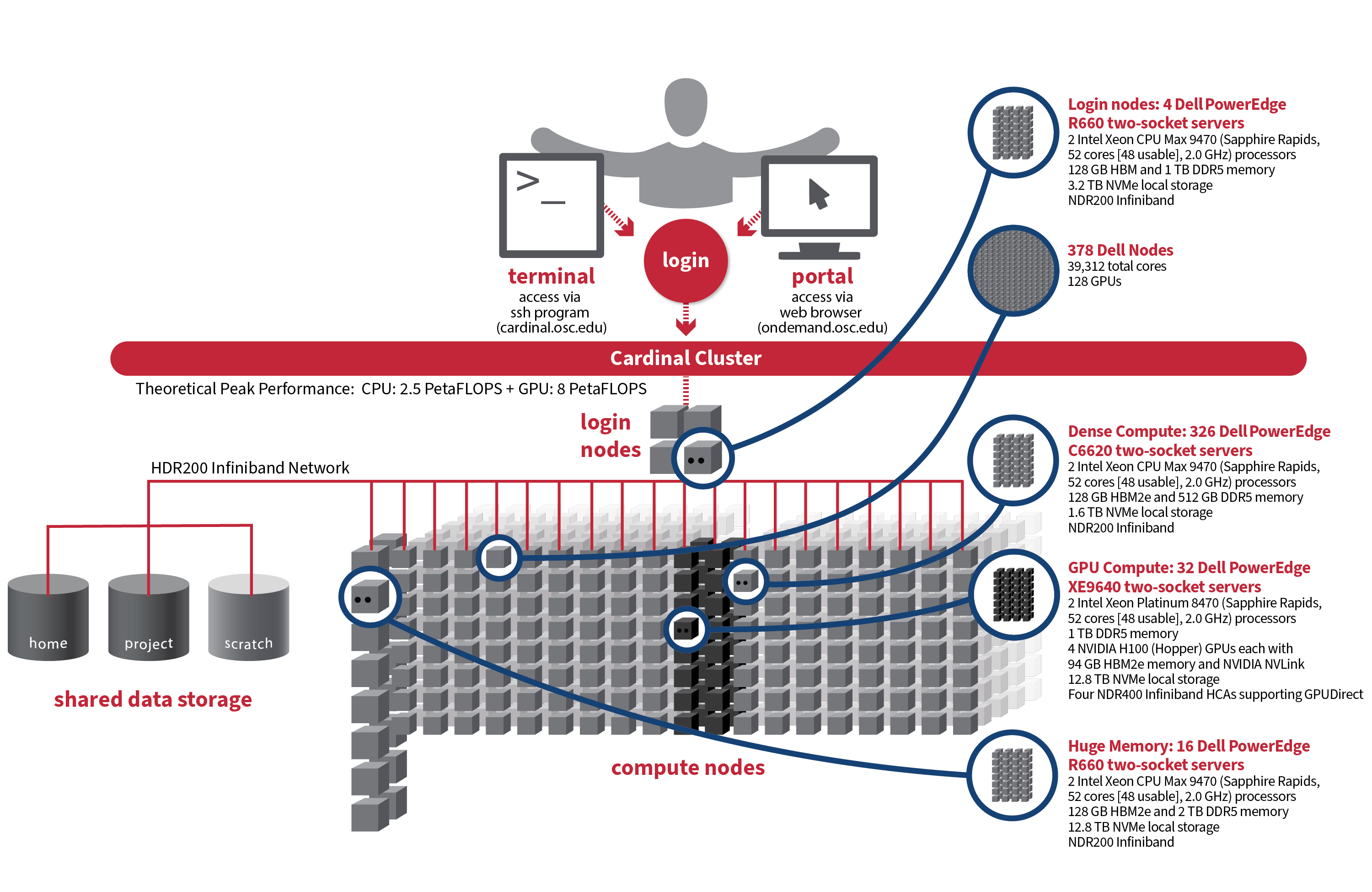
Cardinal
AutoDock
AutoDock is a a suite of automated docking tools. It is designed to predict how small molecules, such as substrates or drug candidates, bind to a receptor of known 3D structure. AutoDock has applications in X-ray crystallography, structure-based drug design, lead optimization, etc.
Nodejs
Nodejs is used to create server-side web applications, and it is perfect for data-intensive applications since it uses an asynchronous, event-driven model
HOWTO: Run Python in Parallel
We can improve performace of python calculation by running python in parallel. In this turtorial we will be making use of the multithreading library to run python code in parallel.
HOWTO: Use GPU in Python
If you plan on using GPUs in tensorflow or pytorch see HOWTO: Use GPU with Tensorflow and PyTorch
This is an exmaple to utilize a GPU to improve performace in our python computations. We will make use of the Numba python library. Numba provides numerious tools to improve perfromace of your python code including GPU support.
Tinker
Tinker is a molecular modeling package. Tinker provides a general set of tools for molecular mechanics and molecular dynamics.
MRIcroGL
MRIcroGL is medical image viewer that allows you to load overlays (e.g. statistical maps), draw regions of interest (e.g. create lesion maps).
Availability and Restrictions
Versions
MRIcroGL is available on Pitzer cluster. These are the versions currently available:
dcm2nii
dcm2niix is designed to convert neuroimaging data from the DICOM format to the NIfTI format. The DICOM format is the standard image format generated by modern medical imaging devices. However, DICOM is very complicated and has been interpreted differently by different vendors. The NIfTI format is popular with scientists, it is very simple and explicit. However, this simplicity also imposes limitations (e.g. it demands equidistant slices).
NVHPC
NVHPC, or NVIDIA HPC SDK, C, C++, and Fortran compilers support GPU acceleration of HPC modeling and simulation applications with standard C++ and Fortran, OpenACC® directives, and CUDA®. GPU-accelerated math libraries maximize performance on common HPC algorithms, and optimized communications libraries enable standards-based multi-GPU and scalable systems programming. Performance profiling and debugging tools simplify porting and optimization of HPC applications, and containerization tools enable easy deployment on-premises or in the cloud.