In this demonstration, we will submit a parallel job to the cluster using R. Most parallelization concepts in R are centered around loop-level parallelism with independence, where each iteration acts as a separate simulation. There are a variety of strategies based on the concept of:
- foreach - enumerating through the contents of a collection
- apply - application of a function to a collection
This is a toy example, hence we use a function that will generate values sampled from a normal distribution and summing the vector of those results; every call to the function is a separate simulation. But sometimes simple is the best way to demonstrate using OSC resources, and to consider a variety of approaches, afforded by the packages that provide foreach, mclapply, and clusterApply. For sufficient parallelism, we generate 100M elements and run this function 100 times.
For reference, this example was heavily adapted from TACC. We then combine several of the strategies in one single R script.
Background: Indirectly, MPI bindings are provided with the Rmpi package, compiled with OpenMPI. On the OSC Owens cluster, this is automatically selected for you if you use the system packages provided with R/3.3.1+.
The code is presented below in 100 lines of parallel_testing.R which you can toggle below.
parallel_testing.R
myProc <- function(size=100000000) {
# Load a large vector
vec <- rnorm(size)
# Now sum the vec values
return(sum(vec))
}
detachDoParallel <- function() {
detach("package:doParallel")
detach("package:foreach")
detach("package:parallel")
detach("package:iterators")
}
max_loop <- 100
# version 1: use mclapply (multicore) - warning - generates zombie processes
library(parallel)
tick <- proc.time()
result <- mclapply(1:max_loop, function(i) myProc(), mc.cores=detectCores())
tock <- proc.time() - tick
cat("\nmclapply/multicore test times using", detectCores(), "cores: \n")
tock
# version 2: use foreach with explicit MPI cluster on one node
library(doParallel, quiet = TRUE)
library(Rmpi)
slaves <- detectCores() - 1
{ sink("/dev/null"); cl_onenode <- makeCluster(slaves, type="MPI"); sink(); } # number of MPI tasks to use
registerDoParallel(cl_onenode)
tick <- proc.time()
result <- foreach(i=1:max_loop, .combine=c) %dopar% {
myProc()
}
tock <- proc.time() - tick
cat("\nforeach w/ Rmpi test times using", slaves, "MPI slaves: \n")
tock
invisible(stopCluster(cl_onenode))
detachDoParallel()
# version 3: use foreach (multicore)
library(doParallel, quiet = TRUE)
cores <- detectCores()
cl <- makeCluster(cores)
registerDoParallel(cl)
tick <- proc.time()
result <- foreach(i=1:max_loop, .combine=c) %dopar% {
myProc()
}
tock <- proc.time() - tick
cat("\nforeach w/ fork times using", cores, "cores: \n")
tock
invisible(stopCluster(cl))
detachDoParallel()
## version 4: use foreach (doSNOW/Rmpi)
library(doParallel, quiet = TRUE)
library(Rmpi)
slaves <- as.numeric(Sys.getenv(c("PBS_NP")))-1
{ sink("/dev/null"); cl <- makeCluster(slaves, type="MPI"); sink(); } # number of MPI tasks to use
registerDoParallel(cl)
tick <- proc.time()
result <- foreach(i=1:max_loop, .combine=c) %dopar% {
myProc()
}
tock <- proc.time() - tick
cat("\nforeach w/ Rmpi test times using", slaves, "MPI slaves: \n")
tock
detachDoParallel() # no need to stop cluster we will use it again
## version 5: use snow backed by Rmpi (cluster already created)
library(Rmpi) # for mpi.*
library(snow) # for clusterExport, clusterApply
#slaves <- as.numeric(Sys.getenv(c("PBS_NP")))-1
clusterExport(cl, list('myProc'))
tick <- proc.time()
result <- clusterApply(cl, 1:max_loop, function(i) myProc())
tock <- proc.time() - tick
cat("\nsnow w/ Rmpi test times using", slaves, "MPI slaves: \n")
tock
invisible(stopCluster(cl))
mpi.quit()
Now, we set up a job to submit R on the cluster, the script again you can toggle below:
owens_job.sh
#!/bin/bash #PBS -l walltime=10:00 #PBS -l nodes=2:ppn=28 #PBS -j oe cd $PBS_O_WORKDIR ml R/3.3.1 # parallel R: submit job with one MPI master mpirun -np 1 R --slave < parallel_testing.R
Then you can submit to the cluster via of qsub owens_job.sh .
If everything is good, you should see some output like:
mclapply/multicore test times using 28 cores: user system elapsed 59.162 1.577 27.099 foreach w/ Rmpi test times using 27 MPI slaves: user system elapsed 12.617 15.204 27.808 foreach w/ fork times using 28 cores: user system elapsed 0.072 0.031 27.003 foreach w/ Rmpi test times using 55 MPI slaves: user system elapsed 8.242 7.230 16.048 snow w/ Rmpi test times using 55 MPI slaves: user system elapsed 7.494 8.052 15.581
This example shows that MPI has the potential to be effectively parallelized on 1 and 2 nodes. In another series, we will look at a more realistic example using more than a single data structure.
Takeaways
When using more than one node use Rmpi or Rmpi SNOW (like in version 4 or version 5). Also, keep in mind that in independent simulation scenarios the use of a do loop means that there are logical processor alignments with the value of max_loop which is set to 100. Because we are setting a fixed problem size (100 times 100M), the benefits of parallelism are chunked by the granularity of the iterations.
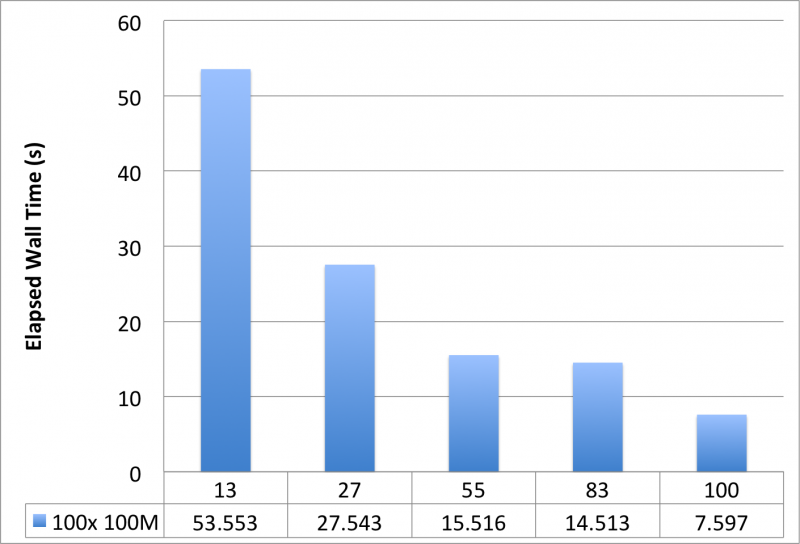
Typically, one sets the max_loop to the number of MPI slaves, or where MPI slaves is a factor of max_loop, because each MPI slave will have its own iteration to perform. However, this all depends on whether or not you have sufficient work on a single iteration. With max_loop set to 100, we are interested in strong scaling effects for a fixed problem size. For this example, the chart below, using 4 nodes of 28 cores (112 cores total available), makes it clear that you should set max_loop equal to MPI slaves.
parallel_testing_version5_only.R
myProc <- function(size=100000000) {
# Load a large vector
vec <- rnorm(size)
# Now sum the vec values
return(sum(vec))
}
max_loop <- 100
## version 5: use snow backed by Rmpi
library(Rmpi) # for mpi.*
library(snow) # for clusterExport, clusterApply
slaves <- as.numeric(Sys.getenv(c("PBS_NP")))-1
{ sink("/dev/null"); cl <- makeCluster(slaves, type="MPI"); sink(); } # MPI tasks to use
clusterExport(cl, list('myProc'))
tick <- proc.time()
result <- clusterApply(cl, 1:max_loop, function(i) myProc())
tock <- proc.time() - tick
cat("\nsnow w/ Rmpi test times using", slaves, "MPI slaves: \n")
tock
stopCluster(cl)
mpi.quit()