Antibiotic resistance causes 23,000 deaths and two million infections each year in the United States, according to the Centers for Disease Control and Prevention. Across the globe, antibiotic resistance is growing, and it is a threat to everything from food production to healthcare to saved lives.
“Bacterial infections are arguably some of the most serious threats to humankind to date,” shared Focco van den Akker, Ph.D., an associate professor of biochemistry at Case Western Reserve University. “The options for treating infections have dwindled substantially, and the reports of antibiotic resistant pathogens are increasing at an alarming rate.”
With the help of the Ohio Supercomputer Center (OSC), van den Akker is trying to unravel the mysteries of certain bacterial enzymes that block medications from working. His discoveries could lead to new ways to treat bacterial infections.
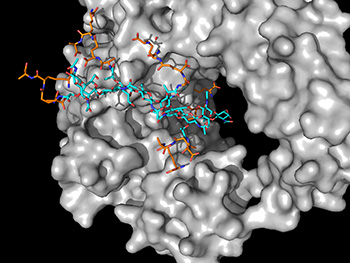
One of his recent studies focuses on the lytic translgycosylase enzyme from a major foodborne pathogen called Campylobacter jejuni. This pathogen is the second highest cause of hospitalization by bacterial infection amongst adults over 65 years old. Using powerful OSC computational and storage resources, insights from van den Akker’s research could lead to new treatments for this pathogen.
“Our computational and experimental results reveal how this important bacterial enzyme works at a molecular level to modify its bacterial cell wall,” van den Akker said. “The bacterial cell wall is a known and current drug target for many antibiotics such as penicillin. Gaining molecular insights into these cell-wall processes could lead to the development of new antibiotics, including new classes of antibiotics, to combat the current antibiotic resistance problem.”
Van den Akker said that gaining insights into how the lytic transglycosylase enzyme functions would lead to a broader understanding of similar enzymes from other bacterial pathogens that stop medications from working.
“Such compounds could lead to new therapeutic opportunities to treat bacterial infections,” he said.
Running Nanoscale Molecular Dynamics (NAMD) software on OSC’s Owens Cluster, van den Akker performed long, intense simulations to perform calculations.
“The length of our simulations was up to one millionth of a second, which for an all-atom simulation is a very long simulation that took one month of calculations using 28 processors at a time,” van den Akker said. “The software and web-based user interface of OSC are about as good as it gets, I was very impressed.”
___________
PROJECT LEAD // Focco van den Akker, Ph.D., Case Western Reserve University
RESEARCH TITLE // Molecular dynamics simulations and inhibitor screening of the lytic transglycosylase protein
FUNDING SOURCE // Case Western Reserve University
WEBSITE // case.edu/med/biochemistry/vandenAkker_lab