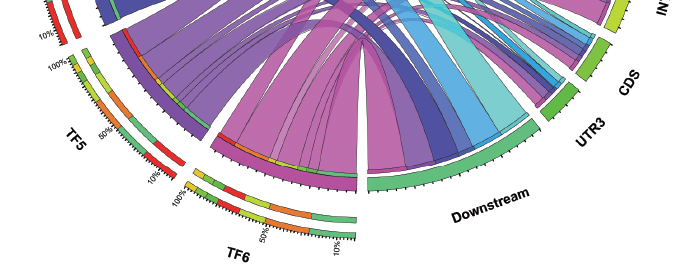
This illustration, developed by Erich Grotewold at The Ohio State University, represents the distribution of binding sites for several transcription factors (TFs 1- 6) in the genome of maize.
Erich Grotewold, a professor of molecular genetics and horticulture and crop science at The Ohio State University, is leveraging the resources of the Ohio Supercomputer Center as part of his studies to address fundamentally important questions in plant research.
He and project co-principle investigators Andrea Doseff, an Ohio State Medical Center and Department of Molecular Genetics associate professor, and John Gray, an associate professor at the University of Toledo, are the recipients of a $4.23 million grant from the National Science Foundation Plant Genome Program to study “Systems Approaches to Identify Gene Regulatory Networks in the Grasses.”
“Establishing gene regulatory networks and linking system components to agronomic traits is an important emerging theme in plant systems biology,” Grotewold said. “This is the first concerted effort to comprehensively dissect the gene regulatory networks that target the metabolism of phenolic compounds, found in maize, other cereal crops and commonly-consumed vegetables.”
These compounds include phenylpropanoids, lignins, flavonoids and hydroxamic acids, all of fundamental agricultural importance. For example, lignin is a complex plant polymer important in biofuel considerations, which is gaining significance as a feedstock for the generation of biochemicals and biomaterials.
As a component of this project, Grotewold is integrating genetics, molecular biology, biochemistry, mathematics, and bioinformatics for the comparative transcriptional genomics of phenolic genes across grass crops.
Specifically, he’s combining the discovery of novel transcription factors that control maize phenolic accumulation with the identification of their interaction partners and of direct target genes for individual and combinations of transcription factors. In molecular biology and genetics, a transcription factor is a protein that binds to specific DNA sequences, thereby controlling the flow of genetic information from DNA to messenger RNA.
“Some of the bioinformatics approaches involve alignment of hundreds of millions of biological sequences to reference genomes, as the first step in the analysis pipeline towards the discovery of the binding regions for often novel transcription factors,” said Grotewold. “Similarly, simulation and modeling approaches will be involved in prediction and inference of gene regulatory networks, which will be validated experimentally. Supercomputers are paramount for this complexity of analysis.”
These findings will accelerate the study of the regulation of other important metabolic and developmental pathways in maize and other grasses. Results are being made widely available through GRASSIUS (grassius.org), a web-accessible knowledge base hosted by the Grotewold lab.
Project Lead: Erich Grotewold, The Ohio State University
Research Title: Gene regulatory networks that target the metabolism of phenolic compounds in maize
Funding Source: National Science Foundation
Website: http://eglab.osu.edu/home